So, if you've followed on from part I, you'll know that I've been looking at packages to visualise KEGG pathway maps. What may not have been clear is why.
I've been working on comparative genomics of a couple of different bacterial genera and - for one of them - in much more detail on constructing models of metabolism for a number of isolates, and combining them into a sort of panmetabolism, analogous to the pangenome. But I needed a way to visualise and represent the similarities and differences between the isolates. KEGG pathways, being curated, standardised, and lovingly rendered in their pathway maps, are a natural choice for this.
So, as an example use case, imagine that I've got about 30 genomes representing six or so species of a single bacterial genus, and that we've been through the sequencing, annotation, and metabolic reconstruction process. Now I want to see where the annotated metabolic genes of my genus sit in the overall metabolic map for KEGG orthologues: ko01100.
 |
| KEGG map ko01100 |
KEGG Orthologues
So, where am I getting my data from? Starting with the features in our bacteria, assume for now that I've got my organism data in a local SQLite3 database because, well, I do. Assume also, because it's true, that I can get all the relevant features and their associated KEGG orthologue IDs in a table called ft_ko. The schema for this table is: |
| ft_ko table schema |
KEGG orthologue identifiers (KOs, format K[0-9]{5}) are the principal way in which gene products are identified in KEGG pathways, as members of manually-defined 'orthologue' groups. These are intended to provide a way of annotating 'equivalent' genes across many organisms in terms of their roles in metabolism, rather than - say - in terms of sequence similarity. Genes/features are assigned KO identifiers during annotation, which associates them with at least one element of at least one KEGG pathway. In this case, our genome's KO identifiers were found using KAAS.
KGML
So, we've given all our features KO IDs to associate them with elements of a KEGG pathway, but how do we get our pathway data? KEGG provides the information in KGML (KEGG Markup Language), a dialect of XML, produced from KEGG's internal 'KGML+'. You can download KGML for each of the organism and orthologue maps at KEGG. However, you can't do this for the 'empty' maps that don't carry organism/orthologue information (e.g. map01100).The KGML data format has been stable for a couple of years, and the specification is given here, with a class diagram for the data structure shown below:
 |
| KGML class diagram |
The other thing to know is that this specification doesn't always correspond to how KEGG presents KGML to you as a user. The class diagram suggests that reactions, substrates and products each generalise an entry, and have corresponding 'required' node IDs. The 'required' ID is not typically present, as this extract from ko00020 shows:
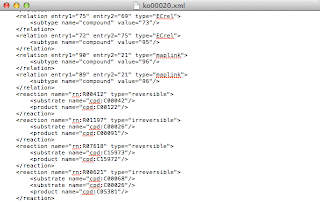 |
| KGML extract from ko00020.xml |
The desirable capability - mentioned above - to be to be able to make modifications to the pathway model (including visualisation choices), and then write those out to KGML, would enable transparent use in a processing pipeline. But since several KGML files from KEGG don't conform to their own specification, my ambition to write strictly valid KGML was tempered to a more pragmatic don't write any KGML that's less compliant than the input. We want, at least, to be able to roundtrip (modulo the XML text formatting).
I like to work in Python, and to use Biopython, but Biopython is short of a KGML parser. There are other KGML parsers out there for Python, such as this one by Giovanni Marco Dall'Olio. He wrote it specifically to represent the pathway as a NetworkX digraph (and thought that this made it unsuitable for Biopython), which is fair enough and makes sense for many uses. It's not been modified for a while, though, and doesn't do exactly what I want, which is capture the graphical information. There's also Eric Xu's similar parser/pathway module as part of kegg-dfba, that has a similar restricted usability. So we need an object model, a parser, and a visualisation too.
It's not always a good idea to assume that the best object model is a slavish clone of either a relational database structure, or DOM tree. In this case, because I wanted to enable relatively transparent use to take in KGML and write out KGML (maybe with a bit of a tweak), I thought that the lazy approach of mimicking the DOM tree was appropriate.
Hey Ronald, what's in the bag? I mean XML?
At first I expected that the KGML file for any pathway would give me enough information to render the pathway as I saw it at KEGG without too much effort. I was wrong.Taking ko01100 (metabolic pathways) as my first example, everything looked promising. Each compound entry element had a graphics subelement describing a circle. All the map entry elements had a roundrectangle graphics subelement. And all the ortholog entry elements, which are associated with and describe reactions, had a line graphics subelement. The ortholog lines lead between the (sometimes multiple) substrate and product compounds. Taken together, these are in fact all the components we need to reproduce the pathway map. The same was true for the ko01110 and ko01120 maps. So far, so good. As long as I could control the display properties of those individual elements, I knew I could get the 'tube map' effect I'm looking for.
But then I took a look at the first module map, ko00010: Glycolysis/gluconeogenesis. Now we have a completely different mode of representation.
 |
| KEGG pathway map for ko00010 |
The packages I covered in part I all handle this issue in the same way: we have the locations of the ortholog, reaction, compound and map elements, and we can infer the directions of any arrows between them from the relation and reaction elements. I thought that this would be easier with a suitable pathway data model, from which we can draw the corresponding arrows as straight lines between the graphics components in each pathway, with arrowheads indicating that the reaction type is either reversible or irreversible. We can draw dashed lines to indicate that a relation type is a maplink, or solid lines to show it has the ECrel type. But we can't easily reproduce the pathway layout in the diagram. I did experiment with arcs and straight lines in early versions of the code, but I wasn't happy with either of those outputs, so I didn't force them on you.
This left me with an obvious option if I wanted to retain KEGG's pathway layout: to take a MapMan-like approach and use the KEGG .png indicated in the pathway element's image field as a backdrop, on which we overlay the ortholog and compound graphical elements. But where do we get these? Well, if you've thought ahead, or have access to the KEGG FTP site, you can just download them en masse. Be warned, though - the .pngs may change over time, so you really want the one that accompanies the KGML file, which means that we would be as well to be able to download the corresponding .png on demand for the fresh KGML, when we need it. So, while we're at it, we might as well provide the ability to grab the KGML on-the-fly, too. Some helper functions to grab data direct from KEGG could be nice, then…
Labelling in KGML diverges from the corresponding pathway map rendering, too. The map graphics element names are the same as we see in the pathway map, but the names for each ortholog in the KGML file are the KO identifiers, rather than EC numbers, and the compound labels are compound identifiers (format cpd:C[0-9]{5}). If we want to relate these to the compound common names, gene names, or EC numbers we need another source of information. Happily, Biopython provides the Bio.KEGG module to help with this.
So, finally, in the next post I'll put up a link to the code and give some examples of use.
No comments:
Post a Comment